Claudins are cell–cell adhesion proteins that form the backbone of tight junctions (TJs) and selectively seal the paracellular spaces in epithelia. Claudins are tetraspan membrane proteins that polymerize in the cell membrane and form a network of strand-like structures that connect to a similar network on neighboring cells.

They form either barriers or channels to control the transport of small molecules across epithelia. Dysregulation of claudin expression or function has been identified in various cancers, kidney diseases, and intestinal diseases, underscoring the therapeutic potential for targeting claudins. Claudins -15 and -2 are cation channel-forming claudins which can be found in the intestinal epithelia, as well as kidney endothelia.
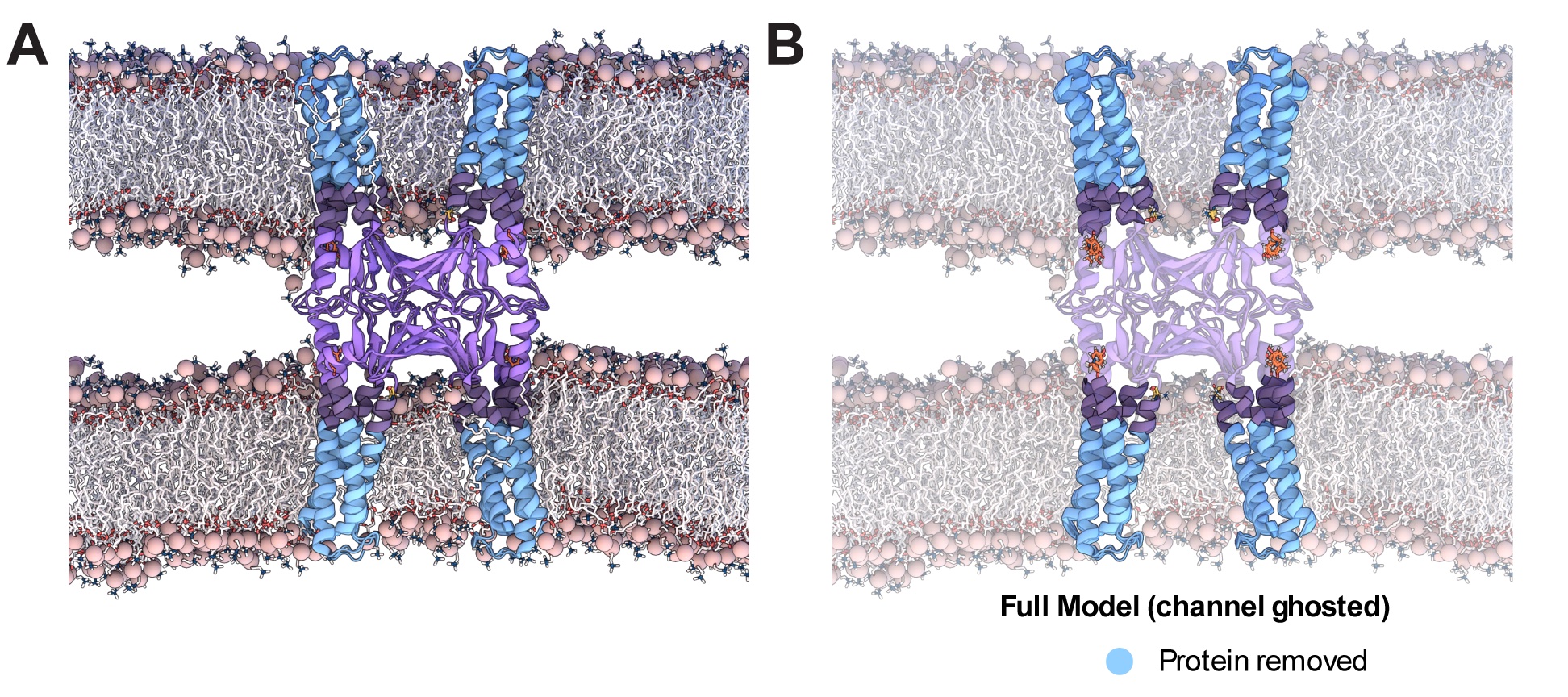
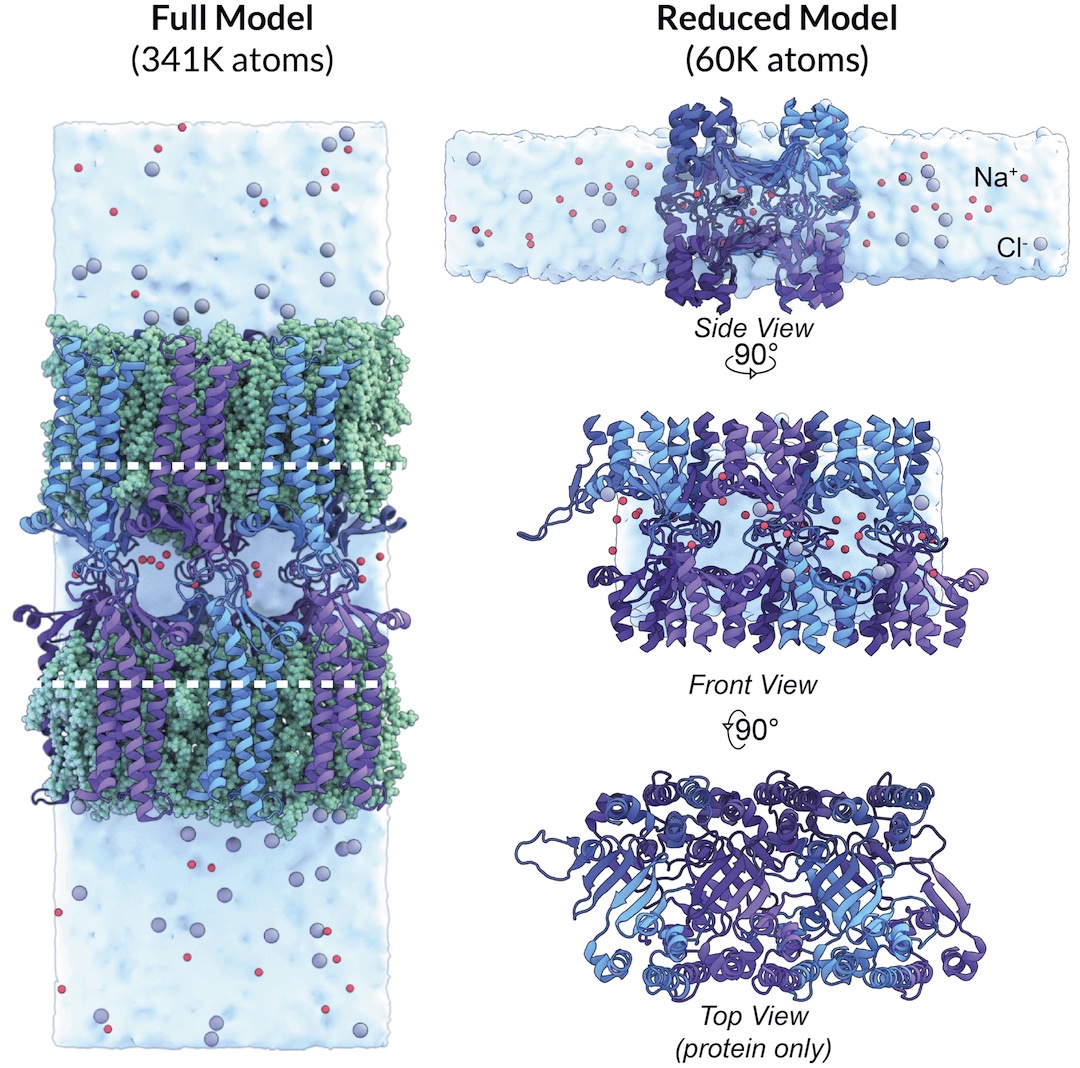
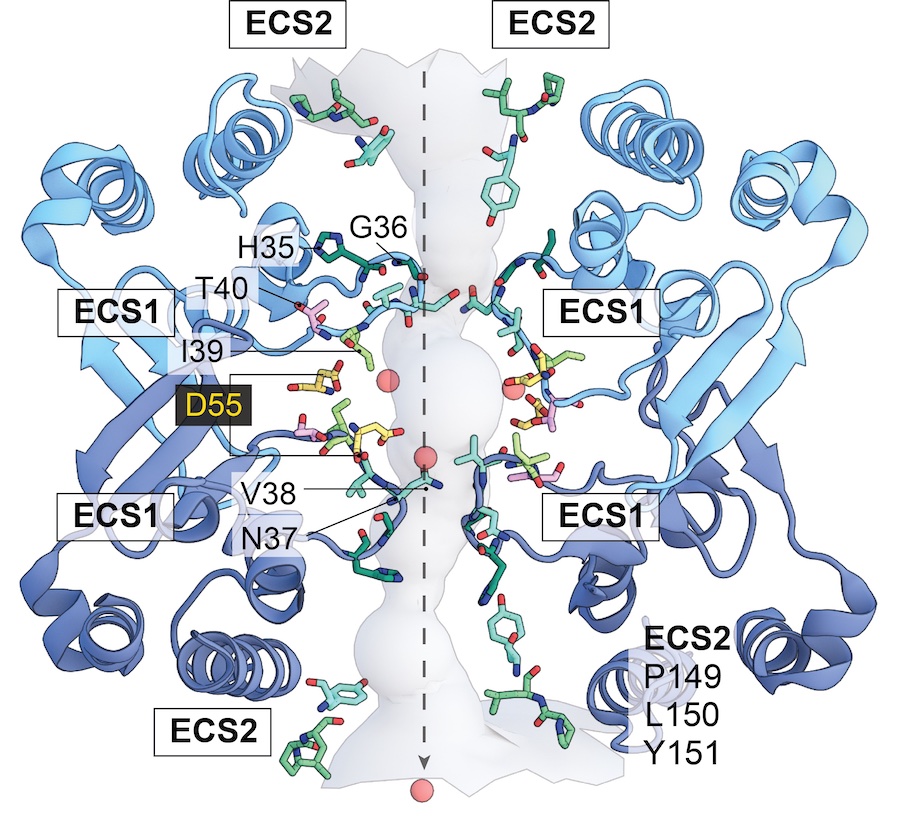
We use Molecular Dynamics (MD) simulations to investigate the paracellular permeability properties of claudin-15 and claudin-2 and to elucidate pore structure and geometry. By applying an electric field to the simulation box, we can simulate ion transport events. We have designed a reduced model of claudin-15 channels by removing the transmembrane helices (blue) and lipid membranes to simulate ion transport in a more computationally efficient manner. This reduced model (purple) allows us to mutate pore-lining residues in order to probe the selectivity properties and three-dimensional arrangement of the pore.